Hands-on tutorial
In this tutorial, you will build a PDI-enabled application step-by-step from a PDI-free base. You will end-up building the C version of the example provided with PDI for the the Trace, Decl'HDF5, and Pycall plugins. Additional examples are available for the other plugins.
Setup
PDI Installation
To run this hands-on tutorial, you will of course first need to install PDI and setup your environment.
On Juwels, PDI is pre-installed as part of the training2022 environment. To load it, just run:
source /p/project/training2022/setup
Setup the hands-on tutorial
Once PDI is installed, you can proceed with getting the sources for the hands-on tutorial from github:
git clone https://github.com/pdidev/tutorial.git
cd tutorial
On Juwels, you can just copy the content from ${PROJECT_training2022}/PDI_tutorial:
cp -R ${PROJECT_training2022}/PDI_tutorial .
cd PDI_tutorial
Compilation
Before compilation, configure the tutorial by detecting all dependencies:
cmake .
For each exercise, once you've modified it, you can compile it by running the following command:
make ex?
Where ? is the number of the exercise.
Execution
In order to speedup the execution on Juwels, it's a good idea to use salloc to reserve one node:
salloc --reservation=parallel-io-day3 -N 1
You can run each exercise with the following command:
srun -n 4 ./ex?
Where ? is the number of the exercise and 4 represents the number of MPI processes to use.
In order to generate clean logs into a file however, the command should be expanded as follow (for example for ex2.):
UCX_LOG_LEVEL=error srun -n 1 -o ex2.juwels.log -e all ./ex2
Now you're ready to work, good luck!
PDI-free code
Ex1. Getting started
Ex1. implements a simple heat equation solver using an explicit forward finite difference scheme parallelized with MPI. The code uses a block domain decomposition where each process holds a 2D block of data.
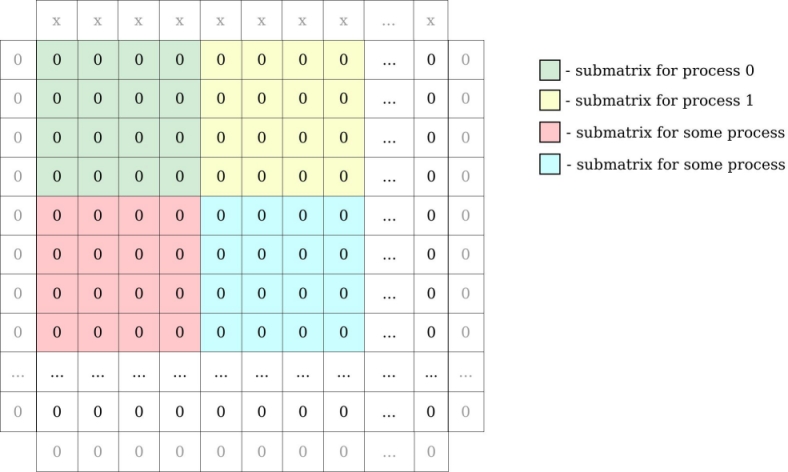
Data domain decomposition in the example

Data domain decomposition in the example

Data domain decomposition in the example

Data domain decomposition in the example
Locally, each process holds its local block of data with one additional element on each side for ghost zones.
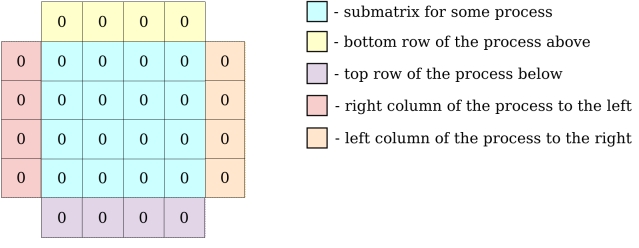
Data domain decomposition in the example

Data domain decomposition in the example

Data domain decomposition in the example

Data domain decomposition in the example
In the following exercises however, PDI will only be used to decouple I/O operations. There is no need to fully dive in the core of the solver described in the PDI example algorithm and implemented in the iter and exchange functions.
The specification tree in the .yml files and the main function are the locations where all the I/O-related aspects will be handled and the only ones you will actually need to fully understand or modify.
- Examine the source code, compile it and run it. There is no input/output operations in the code yet, so you can not see any result.
This example gets its configuration from a file in the YAML format: ex1.yml file. If you're not familiar with YAML, please have a look at our quick YAML format documentation to understand it. The example uses the paraconf library to read this file.
- Play with and understand the code parameters in
ex1.yml. - Set values in
ex1.ymlto be able to run the code with 4 MPI processes.
srun -n 4 ./ex1
PDI core & trace plugin
Ex2. Now with some PDI
Ex2. is the same code as that of ex1. with PDI calls added in main function. In our YAML file (ex2.yml), a new sub-tree has been added under the pdi key. This sub-tree is the PDI specification tree passed to PDI at initialization. Here, the PDI Trace plugin is used to trace PDI calls.
-
Examine the source code, compile it and run it.
-
Add the required
PDI_shareandPDI_reclaimcalls to match the output ofex2.logfile (only the lines matching[Trace-plugin]have been kept). You only need to change theex2.cfile. You can easily check if the files are the same by running the command:diff ex2.log <(grep Trace-plugin ex2.juwels.log)
Decl'HDF5 plugin
Ex3. HDF5 through PDI
In this exercise, the code is the same as in ex2. No need to touch the C code here, modification of the YAML file (ex3.yml) should be enough.
- Examine the YAML file, compile the code and run it.
The Trace plugin (trace) was replaced by the Decl'HDF5 plugin (decl_hdf5) in the specification tree. In its configuration, the dsize variable is written.
-
Write the
psizeandpcoordvariables in addition todsizeto match the content expected as described in theex3.h5dumptext file (use theh5dumpcommand to see the content of your HDF5 output file in the same format as the.h5dumpfile). You can easily check if the files are the same by running the command:diff ex3.h5dump <(h5dump ex3.h5)
To achieve this result, you will need to fill 2 sections in the YAML file.
- The
datasection to indicate to PDI the datatype type of the fields that are exposed. - The
decl_hdf5section for the configuration of the Decl'HDF5 plugin.
ex3.h5 file before, otherwise the data will not be changed.
Ex4. Writing some real data
In this exercise each MPI process will write its local 2D array block contained in the main_field variable to a separate HDF5 file. Once again, this can be done by modifying the YAML file only, no nee to touch the C file.
- Examine the YAML file, compile the code and run it.
Look at the number of blocks, you will have to use the correct number of MPI ranks to run the example.
Notice that in the YAML file, a list was used in the decl_hdf5 section with multiple write blocks instead of a single one as before in order to write to multiple files.
Also notice that this example now runs in parallel with two processes. Therefore it uses "$-expressions" to specify the file names and ensure we do not write to the same file from distinct ranks.
Unlike the other fields manipulated until now, the type of main_field is not fully known, its size is dynamic. By moving other fields in the metadata section, you can reference them from "$-expressions" in the configuration file.
- Use a $-expression to specify the size of
main_field.
Unlike the other fields manipulated until now, main_field is exposed multiple times along execution. In order not to overwrite it every time it is exposed, you need to add a when condition to restrict its output.
- Only write
main_fieldat the second iteration (whenii=1) and match the expected content as described inex4.h5dump.
Ex5. Introducing events
In ex4., two variables were written to ex4-data*.h5, but the file was opened and closed for each and every write. Since Decl'HDF5 only sees the data appear one after the other, it does not keep the file open. Since ii and main_field are shared in an interlaced way, they are both available to PDI at the same time and could be written without opening the file twice. You have to use events for that, you will modify both the C and YAML file.
- Examine the YAML file and source code.
- In the C file, trigger a PDI event named
loopwhen bothiiandmain_fieldare shared. With the Trace plugin, check that the event is indeed triggered at the expected time as described inex5.log(only the lines matching[Trace-plugin]have been kept). - Use the
on_eventmechanism to trigger the write ofiiandmain_field. This mechanism can be combined with awhendirective, in that case the write is only executed when both mechanisms agree. - Also notice the extended syntax that make it possible to write data to a dataset whose name differs from the PDI variable name. Use this mechanism to write
main_fieldat iterations 1 and 2, in two distinct groupsiter1anditer2. Your output should match the content described inex5.h5dump.
Ex6. Simplifying the code
As you can notice, the PDI code is quite redundant. In this exercise, you will use PDI_expose and PDI_multi_expose to simplify the code while keeping the exact same behaviour. For once, there is no need to modify the YAML file here, you only need to modify the C file in this exercise.
- Examine the source code, compile it and run it.
There are lots of matched PDI_share/PDI_reclaim in the code.
- Replace these by
PDI_exposethat is the exact equivalent of aPDI_sharefollowed by a matchingPDI_reclaim.
This replacement is not possible for interlaced PDI_share/PDI_reclaim with events in the middle. This case is however handled by PDI_multi_expose call that exposes all data, then triggers an event and finally does all the reclaim in reverse order.
-
Replace the remaining
PDI_share/PDI_reclaimbyPDI_exposes andPDI_multi_exposes and ensure that your code keeps the exact same behaviour by comparing its trace toex6.log(only the lines matching[Trace-plugin]have been kept). You can easily check if the files are the same by running:diff ex6.log <(grep Trace-plugin ex6.juwels.log)
Ex7. Writing a selection
In this exercise, you will only write a selection of the 2D array in memory excluding ghosts to the HDF5 file. Once again, you only need to modify the YAML file in this exercise, no need to touch the C file.
- Examine the YAML file and compile the code.
As you can notice, now the dataset is independently described in the file.
-
Restrict the selection to the non-ghost part of the array in memory (excluding one element on each side). You should be able to match the expected output described in
ex7.h5dump. You can easily check if the files are the same by running:diff ex7.h5dump <(h5dump ex7-data0x0.h5)
You can achieve this by using the memory_selection directive that specifies the selection of data from memory to write.
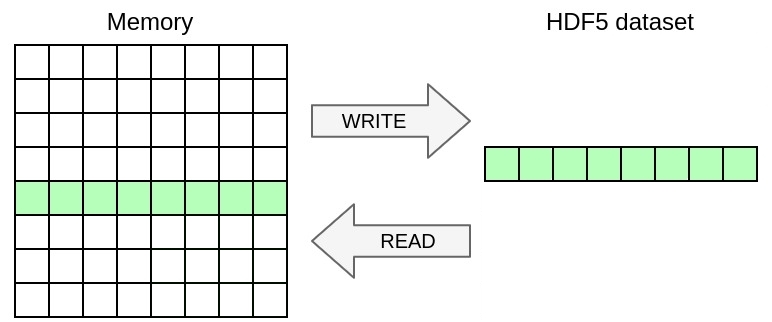
graphical representation

graphical representation

graphical representation

graphical representation
Ex8. Selecting on the dataset size
In this exercise, you will once again change the YAML file to handle a selection in the dataset in addition to the selection in memory from the previous exercise. You will write the 2D array from the previous exercise as a slice of 3D dataset including a dimension for time. Once again, you only need to modify the YAML file in this exercise, no need to touch the C file.
- Examine the YAML file and compile the code.
Notice how the dataset is now extended with an additional dimension for three time-steps.
-
Write the 2D selection from
main_fieldat iterations 1 to 3 inclusive into slices at coordinate 0 to 2 of first dimension of the 3D dataset. Match the expected output described inex8.h5dump. You can easily check if the files are the same by running:diff ex8.h5dump <(h5dump ex8-data0x0.h5)
You can achieve this by using the dataset_selection directive that specifies the selection where to write in the file dataset.
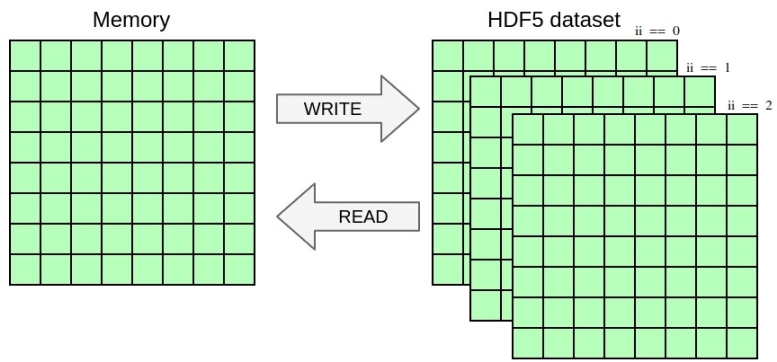
graphical representation

graphical representation

graphical representation

graphical representation
parallel Decl'HDF5
Ex9. Going parallel
Running the code from the previous exercises in parallel should already work and yield one file per process containing the local data block. In this exercise you will write one single file with parallel HDF5 whose content should be independent from the number of processes used. Once again, you only need to modify the YAML file in this exercise, no need to touch the C file.
- Examine the YAML file and compile the code.
The mpi plugin was loaded to make sharing MPI communicators possible.
- Uncomment the
communicatordirective of the Decl'HDF5 plugin to switch to parallel I/O and change the file name so that all processes access the same file. - Set the size of the dataset to take the global (parallel) array size into account. You will need to multiply the local size by the number of processes in each dimension (use
psize). - Ensure the dataset selection of each process does not overlap with the others. You will need to make a selection in the dataset that depends on the global coordinate of the local data block (use
pcoord).
Match the output from ex9.h5dump, that should be independent from the number of processes used. You can easily check if the files are the same by running:
diff ex9.h5dump <(h5dump ex9.h5)
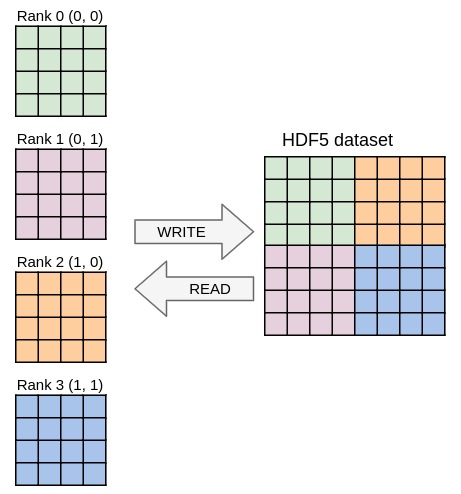
graphical representation of the parallel I\/O

graphical representation of the parallel I\/O

graphical representation of the parallel I\/O

graphical representation of the parallel I\/O
Pycall
Ex10. Post-processing the data in python
In this exercise, you will once again modify the YAML file only and use python to post-process the data in situ before writing it to HDF5. Here, you will write the square root of the raw data to HDF5 instead of the data itself.
- Examine the YAML file and compile the code.
Notice that the Decl'HDF5 configuration was simplified, no memory selection is applied, the when condition disappeared. The dataset name is however explicitly specified now because it does not match the PDI variable name anymore, you will instead write a new variable exposed from python.
The pycall section has been added to load the Pycall plugin. It executes the provided code when the "loop" event is triggered. The with section specifies the variables (parameters) to pass to Python as a set of "$-expressions". The provided code again exposes its result to PDI and multiple blocks can be chained this way.
-
Add the missing parameter to the
withblock to let the Python code process the data exposed inmain_field. -
Modify the Decl'HDF5 configuration to write the new data exposed from Python.
-
Match the output from
ex10.h5dump. You can easily check if the files are the same by running:diff ex10.h5dump <(h5dump ex10.h5)
.py file on the side.
What next ?
In this tutorial, you used the C API of PDI and from YAML, you used the Trace, Decl'HDF5, and Pycall plugins.
If you want to try PDI from another language (Fortran, python, ...) or if you want to experiment with other PDI plugins, have a look at the examples provided with PDI.